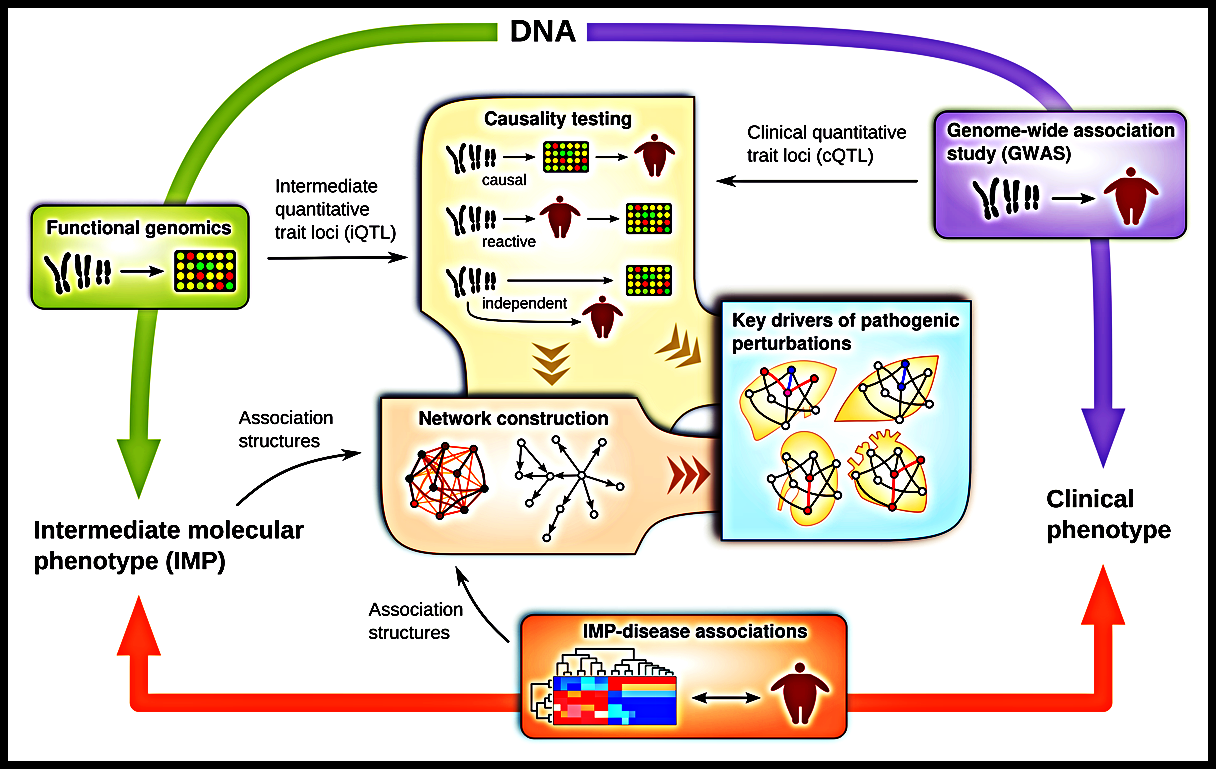
Despite the proliferation of GWAS, the associations found so far have largely failed to account for the known effects of genes on complex disease. Standard approaches also struggle to find combinations of multiple genes that affect disease risk in complex ways.
Now researchers are proposing a new statistical framework that could finally help bioinformatics get off the ground. Or rather, they’ll at least be able to publish more papers as they find more “significant” associations to report.
From the abstract:
We leverage the available population data and optional modeling assumptions, such as Hardy–Weinberg equilibrium (HWE) in the population and linkage equilibrium (LE) between distal loci, to substantially improve power of association and interaction tests. We demonstrate, via simulation and application to actual GWAS data sets, that our approach is substantially more powerful and robust than standard testing approaches that ignore or make naive use of the population sample. We report several novel and credible pairwise interactions, in bipolar disorder, coronary artery disease, Crohn’s disease, and rheumatoid arthritis.